Abstract
Background: Metabolic tumor volume (MTV) has been shown to be a robust prognostic biomarker in diffuse large B cell lymphoma (DLBCL). Currently available semiautomatic software for calculating MTV can be tedious, time consuming and requires manual input from the reader, therefore, limiting its routine application in clinical research. Our objective was to develop a fully automated method for calculating MTV and to validate the algorithm by comparing the automated results with 2 experienced nuclear medicine (NM) readers.
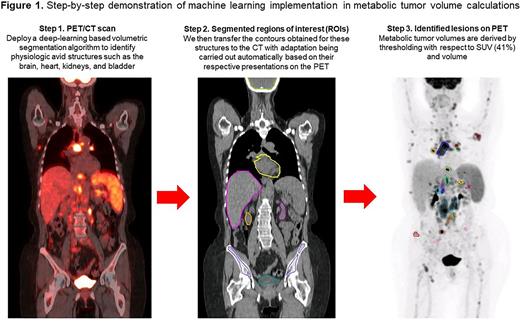
Methods: The automated method designed for this study employed a deep convolutional neural network to segment normal physiologic structures from the CT scans which demonstrate intense avidity on FDG PET scans, including the brain, heart, kidneys, and bladder. The segmentation model was built off the 2D dilated residual U-net using cross entropy and Dice loss as the objective function. The contours obtained for these structures were then automatically transferred and adapted to the PET scans according to their respective PET presentations by aid of an array of ad-hoc image processing algorithms including region growing, active contours, and fast matching. MTV was derived on the PET scans by thresholding with respect to 41% of the maximum SUV within the imaged body volume excluding the above-mentioned normal physiologic structures followed by filtering with a cutoff volume of 1 mL (Figure 1).
The study cohort consisted of 100 patients with newly diagnosed DLBCL who were randomly selected from the Alliance/CALGB 50303 (NCT00118209) trial. De-identified imaging and clinical data were retrieved from The Cancer Imaging Archive. MTV and SUV of the lesion with the highest metabolic activity (SUVmax) were analyzed for the included patients by 2 experienced NM physicians using the Hermes Affinity Viewer and compared to the fully automated results from the developed algorithm.
For examining agreement, we estimated Pearson's correlation coefficients and inter-class correlation coefficients (ICCs) along with corresponding 95% confidence intervals and p-values. For visualization, we displayed scatter plots and Bland-Altman plots between readers and the automated method. All tests were two-sided and statistical significance was considered when p<.05. Statistical software R was used for all statistical analyses.
Results: Among 100 patients included in this final analysis, the mean MTV calculated by reader 1 was 226.47 mL (standard deviation (SD) 260.06 and coefficient of variation (CV) 114.83), for reader 2 was 226.799 mL (SD 261.965 and CV 115.505) and for the automated method (AM) was 205.704 mL (SD 245.825 and CV 119.504). Comparing reader 1 to reader 2, the Pearson's correlation coefficients and ICCs were 0.9997, p<.0001 and 1, p<.0001 (95%CI=1 to 1) for MTV and 1, p<.0001 and 1, p<.0001 (95%CI=1 to 1) for SUVmax, respectively. Comparing reader 1 to AM, the Pearson's correlation coefficients and ICCs were 0.9814, p<.0001 and 0.98, p<.0001 (95%CI=0.96 to 0.99) for MTV and 0.9868, p<.0001 and 1, p<.0001 (95%CI=0.99 to 1) for SUVmax, respectively. Comparing reader 2 to AM, the Pearson's correlation coefficients and ICCs were 0.9818, p<.0001 and 0.98, p<.0001 (95%CI=0.96 to 0.99) for MTV and 0.9868, p<.0001 and 1, p<.0001 (95%CI=0.99 to 1) for SUVmax, respectively. The Bland-Altman plots showed only relatively small systematic errors between the proposed method and the manual readings across the entire data range being examined for both MTV and SUVmax.
Limitations of the automated segmentation algorithm occurred when tumor activity was located in close proximity to normal physiologic structures such as the bladder or kidneys or when normal anatomy had been distorted either due to the disease process or image artifacts such as misregistration or patient motion. Overall, the AM enabled faster MTV calculations (median time 5 minutes) compared to a semiautomatic approach (median time 20 minutes).
Conclusion: The proposed automated method for calculating MTV demonstrates a high agreement with 2 experienced NM readers. Furthermore, the algorithm was highly accurate in classifying FDG-avidity in patients from a multicenter clinical trial involving 17 centers that obtained images on different scanner models with variable reconstruction settings. This approach possess the potential to integrate PET-based biomarkers in clinical trials.
Disclosures
Alderuccio:Pyramid: Consultancy; ADC Therapeutics: Consultancy, Research Funding; Agios: Consultancy. Lossos:LRF: Membership on an entity's Board of Directors or advisory committees; NCI: Research Funding; Adaptive: Honoraria. Moskowitz:Merck: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal